
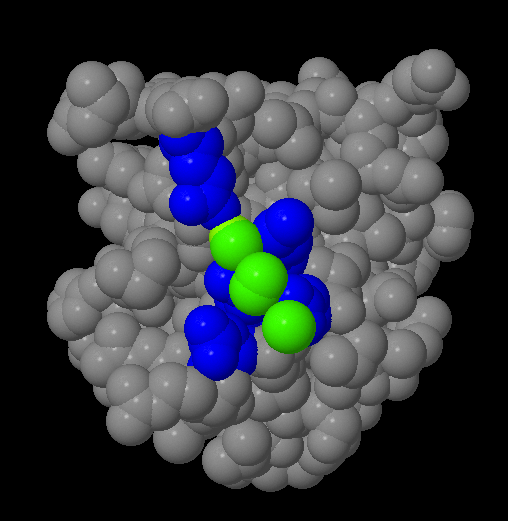
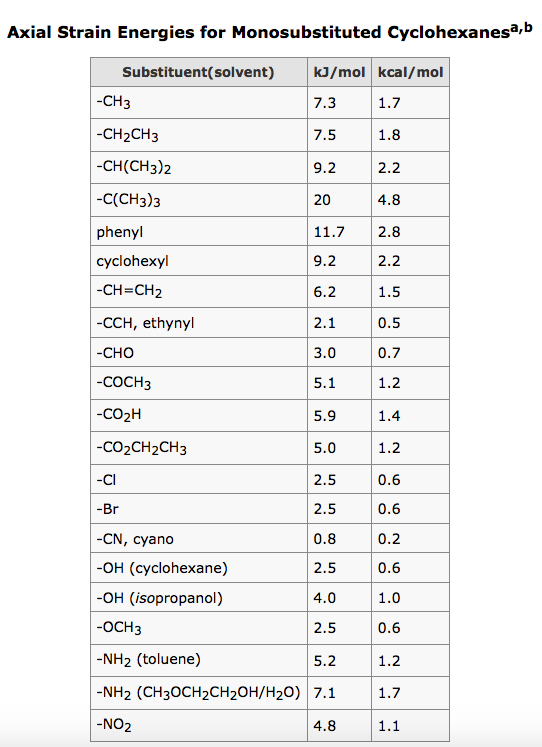
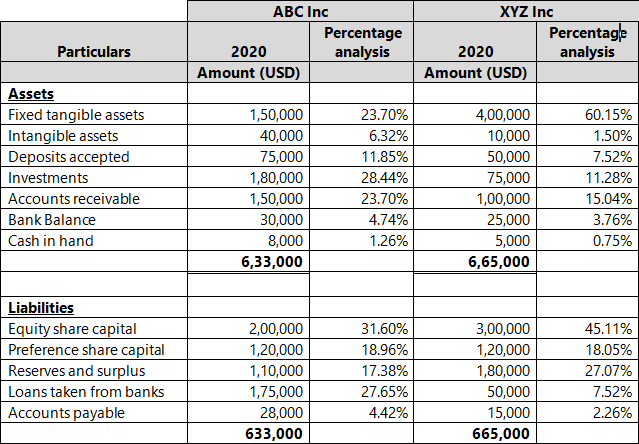
I am attempting to use charges (or temperature) purely to discriminateĪtoms of the same element by coordination (here: sp2, sp2+x, sp3), as Whereas Rasmol (and, I do believe Xmol) is fine with pretty much any In Jmol, however, only integers are recognized (as formal charges), I like the XYZ format for its simplicity andįlexibility in optionally conveying auxiliary scalars and vectors. Open the molden.inp file with molden and write out a new file (select "write" in the moden menu).I just re-discovered Jmol and the amazing progress made by theĬoming from Rasmol, I ran into a Jmol peculiarity reading partialĬharges from XYZ files. DALTON: The molden file from dalton cannot be opened by jmol."o2_state_tpssd3_prod_oh_opt_cosmo.xyz" "o2_state_tpssd3_o_opt_cosmo.xyz"Įcho "load structure.xyz spacefill 0.2" > orbital_$i".spt"Įcho 'isosurface sign "'$i.cube'" translucent 0.4' > orbital_$i".spt"Įcho "write jpg $i.jpg" > orbital_$i".spt" "o2_state_tpssd3_react_oo_opt_cosmo.xyz" "o2_state_tpssd3_oh2_opt_cosmo.xyz" "o2_state_tpssd3_react_oh_opt_cosmo_singlet.xyz" "o2_state_tpssd3_react_ooh_opt_cosmo_singlet.xyz" Load FILES "o2_state_tpssd3_ooh2_opt_cosmo.xyz" The following java script can be inserted in the Jmol scripting window - It loads the files rotate them (adjust this according to your needs), writes the distances between atoms 49/50, 49/1, 49/10 and 49/30 and prints a frame in jpg format.Script to generate many MOs for i in do cubegen 0 MO="$i" orbitals.fchk "$i".cube done Jmol structures and molecular orbitals Note for an unrestricted calculation, use "AMO" or "BMO" instead. Use the utility program cubegen cubegen 0 MO= gaussian.fchk.Note: make sure the gaussian module is loaded (aurora: module load gaussian/16.B.01-AVX2) Transform the ".chk" with the utility program formchk gaussian.chk gaussian.fchk.Run a gaussian wave function optimization and make sure you obtain the ".chk" file, e.g.We focus here on generation of cube files that can be visualized with Jmol (see below). In gaussian, the can be generated in different ways and visualized with Jmol (see below). They are particularly useful for interpreting electronic spectroscopy (i.e. Molecular orbitals are often useful for understanding the electronic structure of a molecule. xyz or pdb file can be written out with from File => Save Coordinates.IMPORTANT: The align will happend according to the "top" molecule (marked with "T" in the VMD main window - this molecule will have unchanged coordinates) It only works if there are equally many of the atom types in the fwo files!). (note that they should have the same name in the pdb/xyz files that are compared. Now type for the atoms that should be aligned (in box upper left) name ATOM1 ATOM2 ATOM3 etc.Check that it is the right molecules that are read in (try "Add all"). Extensions => Analysis => RMSD Trajectory Tool.Note: this can also be done (often faster) with Maestro "Quick Align" Type vmd > set sel4 įor all examples, write the pdb using writepdb selection: vmd > $selX writepdb pdb_out.pdb.Select whole molecules, here waters - and only waters Save pdb with selection of atoms within 5 Å of another atom, here "Mn" type command set sel1 Ĭheck that the number of atoms fit by command $sel1numĮxample 2.Saving pdbs with different selections (examples) within 5 of name FE (selects all atoms withn 5 Å of FE).name CA (typically selects alpha-C in protiens if the "normal" pdb name for alpha carbons are used).Graphics => Representations: The menu "Selected Atoms" can be used.Read in pdb or xyz structure: vmd pdb_name.pdb or vmd xyz_name.xyzĪfter starting a VMD shell (see above) go to.VMD can be found here: starting vmd and loading structures and Schulten, K., "VMD - Visual Molecular Dynamics", J.
#Jmol xyz example software#
Useful commands and features of the powerfull VMD software (Humphrey, W., Dalke, A. Visualization tools for quantum chemistry


 0 kommentar(er)
0 kommentar(er)
